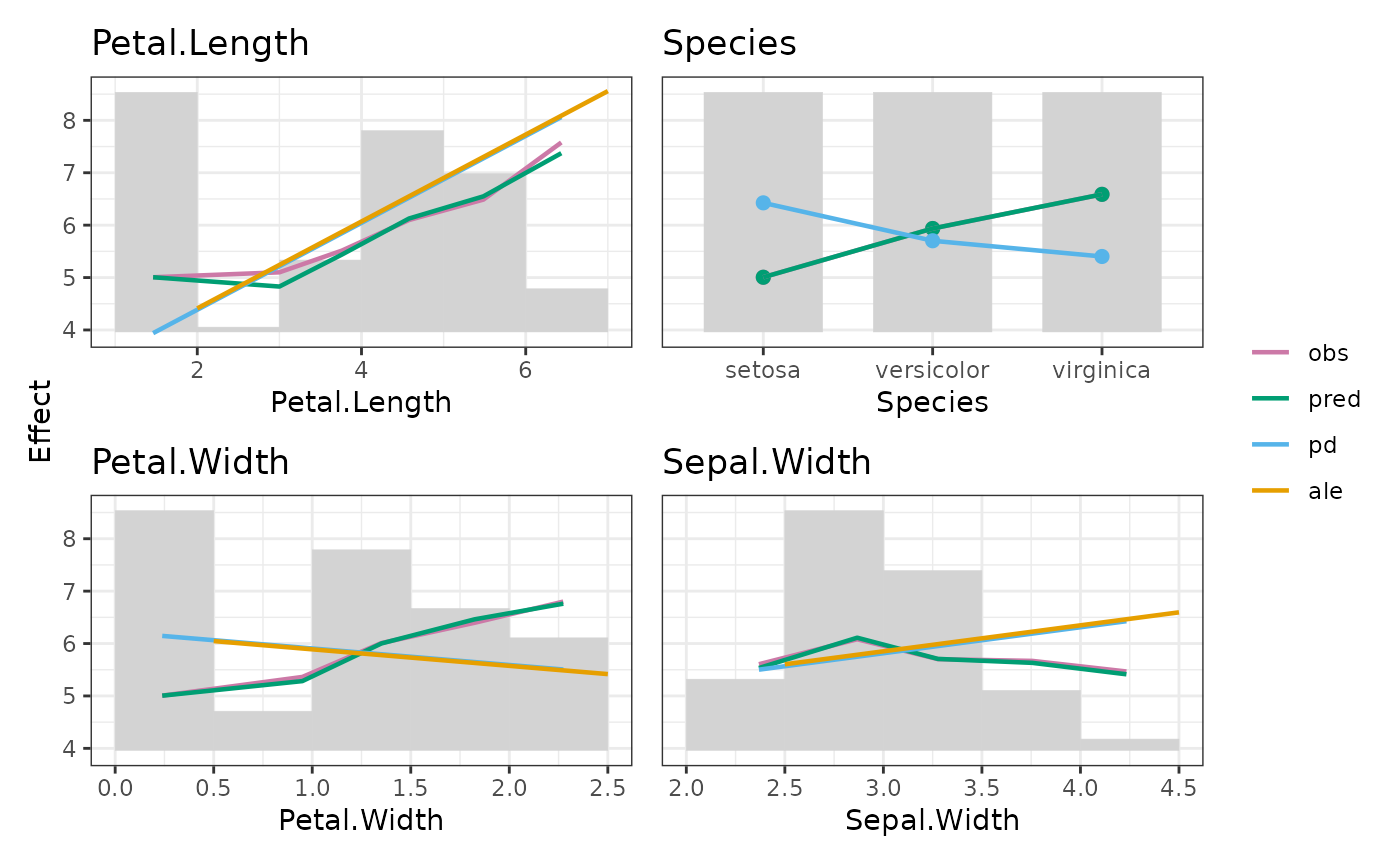
This is the main function of the package. By default, it calculates the following statistics per feature X over values/bins:
"y_mean": Average observed
yvalues. Used to assess descriptive associations between response and features."pred_mean": Average predictions. Corresponds to "M Plots" (from "marginal") in Apley (2020). Shows the combined effect of X and other (correlated) features. The difference to average observed y values shows model bias.
"resid_mean": Average residuals. Calculated when both
yand predictions are available. Useful to study model bias."pd": Partial dependence (Friedman, 2001): See
partial_dependence(). Evaluated at bin averages, not at bin midpoints."ale": Accumulated local effects (Apley, 2020): See
ale(). Only for continuous features.
Additionally, corresponding counts/weights are calculated, and standard deviations of observed y and residuals.
Numeric features with more than discrete_m = 13 disjoint values are binned via
breaks. If breaks is a single integer or "Sturges", the total bin range is
calculated without values outside +-2 IQR from the quartiles.
Values outside the bin range are placed in the outermost bins. Note that
at most 9997 observations are used to calculate quartiles and IQR.
All averages and standard deviation are weighted by optional weights w.
If you need only one specific statistic, you can use the simplified APIs of
feature_effects(object, ...)
# Default S3 method
feature_effects(
object,
v,
data,
y = NULL,
pred = NULL,
pred_fun = stats::predict,
trafo = NULL,
which_pred = NULL,
w = NULL,
breaks = "Sturges",
right = TRUE,
discrete_m = 13L,
outlier_iqr = 2,
calc_pred = TRUE,
pd_n = 500L,
ale_n = 50000L,
ale_bin_size = 200L,
seed = NULL,
...
)
# S3 method for class 'ranger'
feature_effects(
object,
v,
data,
y = NULL,
pred = NULL,
pred_fun = NULL,
trafo = NULL,
which_pred = NULL,
w = NULL,
breaks = "Sturges",
right = TRUE,
discrete_m = 13L,
outlier_iqr = 2,
calc_pred = TRUE,
pd_n = 500L,
ale_n = 50000L,
ale_bin_size = 200L,
...
)
# S3 method for class 'explainer'
feature_effects(
object,
v = colnames(data),
data = object$data,
y = object$y,
pred = NULL,
pred_fun = object$predict_function,
trafo = NULL,
which_pred = NULL,
w = object$weights,
breaks = "Sturges",
right = TRUE,
discrete_m = 13L,
outlier_iqr = 2,
calc_pred = TRUE,
pd_n = 500L,
ale_n = 50000L,
ale_bin_size = 200L,
...
)
# S3 method for class 'H2OModel'
feature_effects(
object,
data,
v = object@parameters$x,
y = NULL,
pred = NULL,
pred_fun = NULL,
trafo = NULL,
which_pred = NULL,
w = object@parameters$weights_column$column_name,
breaks = "Sturges",
right = TRUE,
discrete_m = 13L,
outlier_iqr = 2,
calc_pred = TRUE,
pd_n = 500L,
ale_n = 50000L,
ale_bin_size = 200L,
...
)Arguments
- object
Fitted model.
- ...
Further arguments passed to
pred_fun(), e.g.,type = "response"in aglm()or (typically)prob = TRUEin classification models.- v
Variable names to calculate statistics for.
- data
Matrix or data.frame.
- y
Numeric vector with observed values of the response. Can also be a column name in
data. Omitted ifNULL(default).- pred
Pre-computed predictions (as from
predict()/pred_fun()). IfNULL, it is calculated aspred_fun(object, data, ...)`.- pred_fun
Prediction function, by default
stats::predict. The function takes three arguments (names irrelevant):object,data, and....- trafo
How should predictions be transformed? A function or
NULL(default). Examples arelog(to switch to link scale) orexp(to switch from link scale to the original scale). Applied afterwhich_pred.- which_pred
If the predictions are multivariate: which column to pick (integer or column name). By default
NULL(picks last column). Applied beforetrafo.- w
Optional vector with case weights. Can also be a column name in
data. Having observations with non-positive weight is equivalent to excluding them.- breaks
An integer, vector, or "Sturges" (the default) used to determine bin breaks of continuous features. Values outside the total bin range are placed in the outmost bins. To allow varying values of
breaksacross features,breakscan be a list of the same length asv, or a named list with breaks for certain variables.- right
Should bins be right-closed? The default is
TRUE. Vectorized overv. Only relevant for continuous features.- discrete_m
Numeric features with up to this number of unique values should not be binned but rather treated as discrete. The default is 13. Vectorized over
v.- outlier_iqr
If
breaksis an integer or "Sturges", the breaks of a continuous feature are calculated without taking into account feature values outside quartiles +-outlier_iqr* IQR (where <= 9997 values are used to calculate the quartiles). To let the breaks cover the full data range, setoutlier_iqrto 0 orInf. Vectorized overv.- calc_pred
Should predictions be calculated? Default is
TRUE. Only relevant ifpred = NULL.- pd_n
Size of the data used for calculating partial dependence. The default is 500. For larger
data(andw),pd_nrows are randomly sampled. Each variable specified byvuses the same sample. Set to 0 to omit PD calculations.- ale_n
Size of the data used for calculating ALE. The default is 50000. For larger
data(andw),ale_nrows are randomly sampled. Each variable specified byvuses the same sample. Set to 0 to omit ALE calculations.- ale_bin_size
Maximal number of observations used per bin for ALE calculations. If there are more observations in a bin,
ale_bin_sizeindices are randomly sampled. The default is 200. Applied after sampling regardingale_n.- seed
Optional integer random seed used for:
Partial dependence: select background data if
n > pd_n.ALE: select background data if
n > ale_n, and for bins >ale_bin_size.Calculating breaks: The bin range is determined without values outside quartiles +- 2 IQR using a sample of <= 9997 observations to calculate quartiles.
Value
A list (of class "EffectData") with a data.frame per feature having columns:
bin_mid: Bin mid points. In the plots, the bars are centered around these.bin_width: Absolute width of the bin. In the plots, these equal the bar widths.bin_mean: For continuous features, the (possibly weighted) average feature value within bin. For discrete features equivalent tobin_mid.N: The number of observations within bin.weight: The weight sum within bin. Whenw = NULL, equivalent toN.Different statistics, depending on the function call.
Use single bracket subsetting to select part of the output. Note that each data.frame contains an attribute "discrete" with the information whether the feature is discrete or continuous. This attribute might be lost when you manually modify the data.frames.
Methods (by class)
feature_effects(default): Default method.feature_effects(ranger): Method for ranger models.feature_effects(explainer): Method for DALEX explainer.feature_effects(H2OModel): Method for H2O models.
References
Molnar, Christoph. 2019. Interpretable Machine Learning: A Guide for Making Black Box Models Explainable. https://christophm.github.io/interpretable-ml-book/.
Friedman, Jerome H. 2001, Greedy Function Approximation: A Gradient Boosting Machine. Annals of Statistics 29 (5): 1189-1232. doi:10.1214/aos/1013203451.3.
Apley, Daniel W., and Jingyu Zhu. 2016. Visualizing the Effects of Predictor Variables in Black Box Supervised Learning Models. Journal of the Royal Statistical Society Series B: Statistical Methodology, 82 (4): 1059–1086. doi:10.1111/rssb.12377.
See also
Examples
fit <- lm(Sepal.Length ~ ., data = iris)
xvars <- colnames(iris)[2:5]
M <- feature_effects(fit, v = xvars, data = iris, y = "Sepal.Length", breaks = 5)
M
#> 'EffectData' object of length 4, starting with 'Sepal.Width':
#>
#> bin_mid bin_width bin_mean N weight pred_mean y_mean resid_mean
#> 1 2.25 0.5 2.368421 19 19 5.532860 5.605263 0.072403270
#> 2 2.75 0.5 2.867188 64 64 6.111200 6.081250 -0.029950246
#> 3 3.25 0.5 3.277083 48 48 5.706627 5.702083 -0.004544115
#> 4 3.75 0.5 3.756250 16 16 5.631317 5.668750 0.037432757
#> 5 4.25 0.5 4.233333 3 3 5.413218 5.466667 0.053449004
#> y_sd resid_sd pd ale
#> 1 0.6041281 0.2706373 5.501709 5.603654
#> 2 0.7766176 0.3154448 5.749042 5.851598
#> 3 0.8704339 0.3110132 5.952305 6.099543
#> 4 1.0097648 0.2733961 6.189918 6.347487
#> 5 0.2516611 0.2374817 6.426499 6.595432
M |>
update(sort = "pd") |>
plot(share_y = "all")