Identifies outliers in new data based on previously fitted "outForest" object.
The result of predict() is again an object of class "outForest".
All its methods can be applied to it.
Arguments
- object
An object of class "outForest".
- newdata
A new
data.frameto be assessed for numeric outliers.- replace
Should outliers be replaced via predictive mean matching "pmm" (default), by "predictions", or by
NA("NA"). Use "no" to keep outliers as they are.- pmm.k
For
replace = "pmm", from how many nearest OOB prediction neighbours (from the original non-outliers) to sample?- threshold
Threshold above which an outlier score is considered an outlier. The default is 3.
- max_n_outliers
Maximal number of outliers to identify. Will be used in combination with
thresholdandmax_prop_outliers.- max_prop_outliers
Maximal relative count of outliers. Will be used in combination with
thresholdandmax_n_outliers.- seed
Integer random seed.
- ...
Further arguments passed from other methods.
Value
An object of class "outForest".
See also
Examples
(out <- outForest(iris, allow_predictions = TRUE))
#>
#> Outlier identification by random forests
#>
#> Variables to check: Sepal.Length, Sepal.Width, Petal.Length, Petal.Width
#> Variables used to check: Sepal.Length, Sepal.Width, Petal.Length, Petal.Width, Species
#>
#> Checking: Sepal.Length Sepal.Width Petal.Length Petal.Width
#> I am an object of class(es) outForest and list
#>
#> The following number of outliers have been identified:
#>
#> Number of outliers
#> Sepal.Length 1
#> Sepal.Width 1
#> Petal.Length 2
#> Petal.Width 2
iris1 <- iris[1, ]
iris1$Sepal.Length <- -1
pred <- predict(out, newdata = iris1)
outliers(pred)
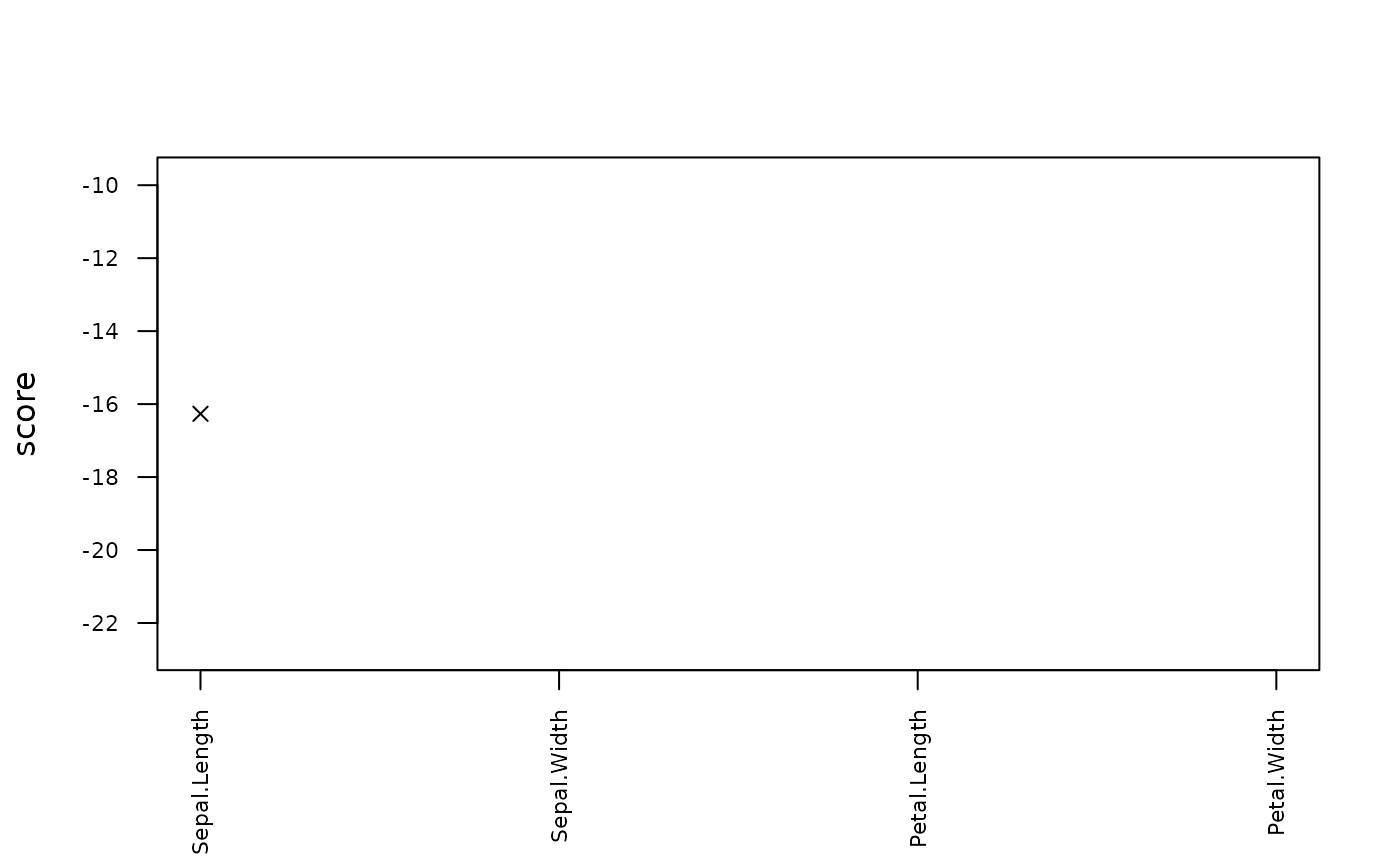
#> row col observed predicted rmse score threshold replacement
#> 1 1 Sepal.Length -1 5.004353 0.3691353 -16.26599 3 5
Data(pred)
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> 1 5 3.5 1.4 0.2 setosa
plot(pred)
 plot(pred, what = "scores")
plot(pred, what = "scores")