Calculates different types of 2D-profiles across two variables. By default, partial dependence profiles are calculated (see Friedman). Other options are response, predicted values, and residuals. The results are aggregated by (weighted) means.
light_profile2d(x, ...)
# Default S3 method
light_profile2d(x, ...)
# S3 method for class 'flashlight'
light_profile2d(
x,
v = NULL,
data = NULL,
by = x$by,
type = c("partial dependence", "predicted", "response", "residual", "shap"),
breaks = NULL,
n_bins = 11L,
cut_type = "equal",
use_linkinv = TRUE,
counts = TRUE,
counts_weighted = FALSE,
pd_evaluate_at = NULL,
pd_grid = NULL,
pd_indices = NULL,
pd_n_max = 1000L,
pd_seed = NULL,
...
)
# S3 method for class 'multiflashlight'
light_profile2d(
x,
v = NULL,
data = NULL,
type = c("partial dependence", "predicted", "response", "residual", "shap"),
breaks = NULL,
n_bins = 11L,
cut_type = "equal",
pd_evaluate_at = NULL,
pd_grid = NULL,
...
)Arguments
- x
An object of class "flashlight" or "multiflashlight".
- ...
Further arguments passed to
formatC()in forming the cut breaks of thevvariables. Not relevant for partial dependence profiles.- v
A vector of exactly two variable names to be profiled.
- data
An optional
data.frame.- by
An optional vector of column names used to additionally group the results.
- type
Type of the profile: Either "partial dependence", "predicted", "response", or "residual".
- breaks
Named list of cut breaks specifying how to bin one or more numeric variables. Used to overwrite automatic binning via
n_binsandcut_type. Ignored for non-numericv.- n_bins
Approximate number of unique values to evaluate for numeric
v. Can be an unnamed vector of length 2 to distinguish between v.- cut_type
Should numeric
vbe cut into "equal" or "quantile" bins? Can be an unnamed vector of length 2 to distinguish between v.- use_linkinv
Should retransformation function be applied? Default is
TRUE.- counts
Should observation counts be added?
- counts_weighted
If
countsis TRUE: Should counts be weighted by the case weights? IfTRUE, the sum ofwis returned by group.- pd_evaluate_at
An named list of evaluation points for one or more variables. Only relevant for type = "partial dependence".
- pd_grid
An evaluation
data.framewith exactly two columns, e.g., generated byexpand.grid(). Only used for type = "partial dependence". Offers maximal flexibility.- pd_indices
A vector of row numbers to consider in calculating partial dependence profiles. Only used for type = "partial dependence".
- pd_n_max
Maximum number of ICE profiles to calculate (will be randomly picked from
data). Only used for type = "partial dependence".- pd_seed
Integer random seed used to select ICE profiles. Only used for type = "partial dependence".
Value
An object of class "light_profile2d" with the following elements:
dataA tibble containing results.byNames of group by variables.vThe two variable names evaluated.typeSame as inputtype. For information only.
Details
Different binning options are available, see arguments below.
For high resolution partial dependence plots, it might be necessary to specify
breaks, pd_evaluate_at or pd_grid in order to avoid empty parts
in the plot. A high value of n_bins might not have the desired effect as it
internally capped at the number of distinct values of a variable.
For partial dependence and prediction profiles, "model", "predict_function", "linkinv" and "data" are required. For response profiles it is "y", "linkinv" and "data". "data" can also be passed on the fly.
Methods (by class)
light_profile2d(default): Default method not implemented yet.light_profile2d(flashlight): 2D profiles for flashlight.light_profile2d(multiflashlight): 2D profiles for multiflashlight.
References
Friedman J. H. (2001). Greedy function approximation: A gradient boosting machine. The Annals of Statistics, 29:1189–1232.
See also
Examples
fit_part <- lm(Sepal.Length ~ Species + Petal.Length, data = iris)
fl_part <- flashlight(
model = fit_part, label = "part", data = iris, y = "Sepal.Length"
)
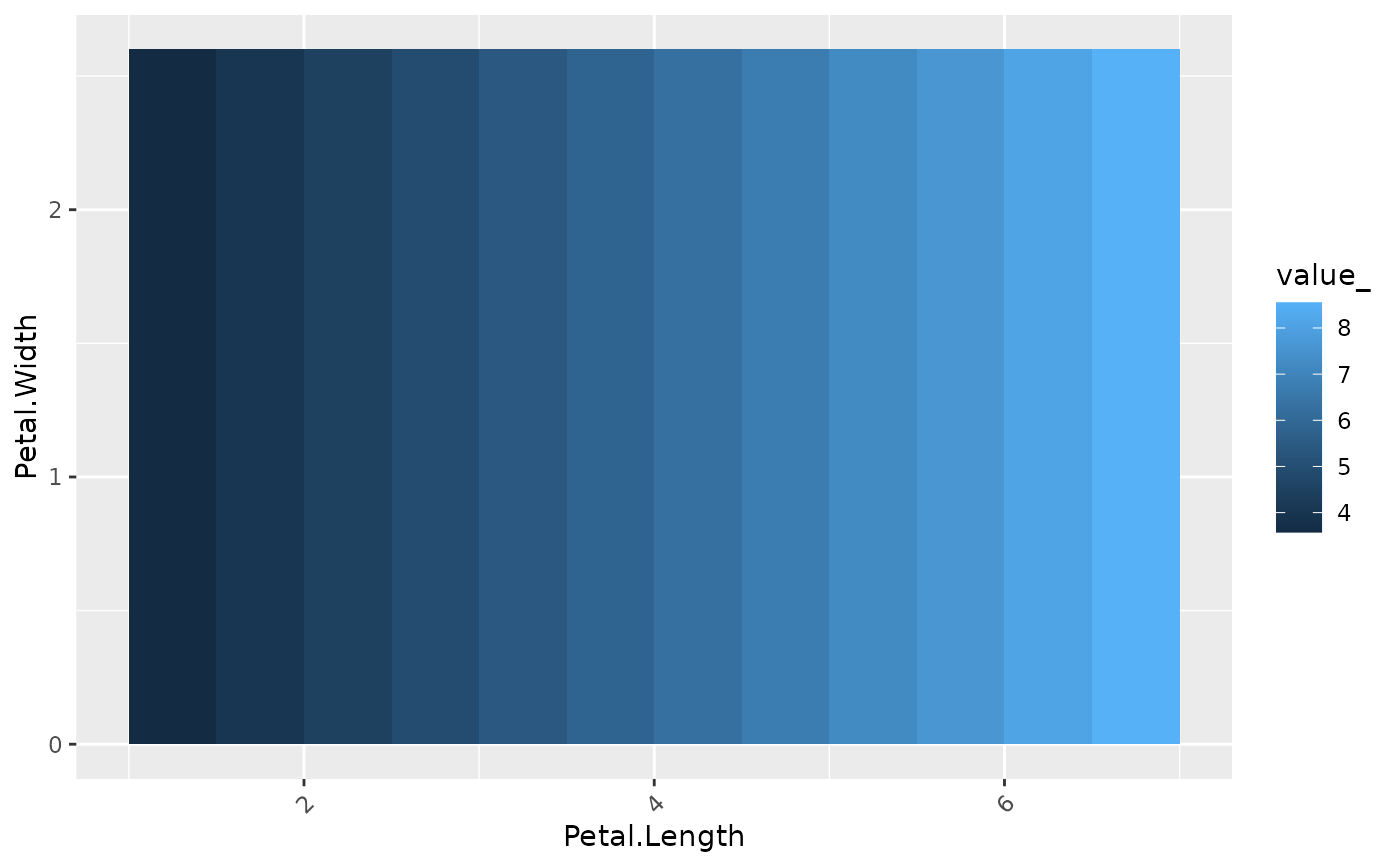
# No effect of Petal.Width
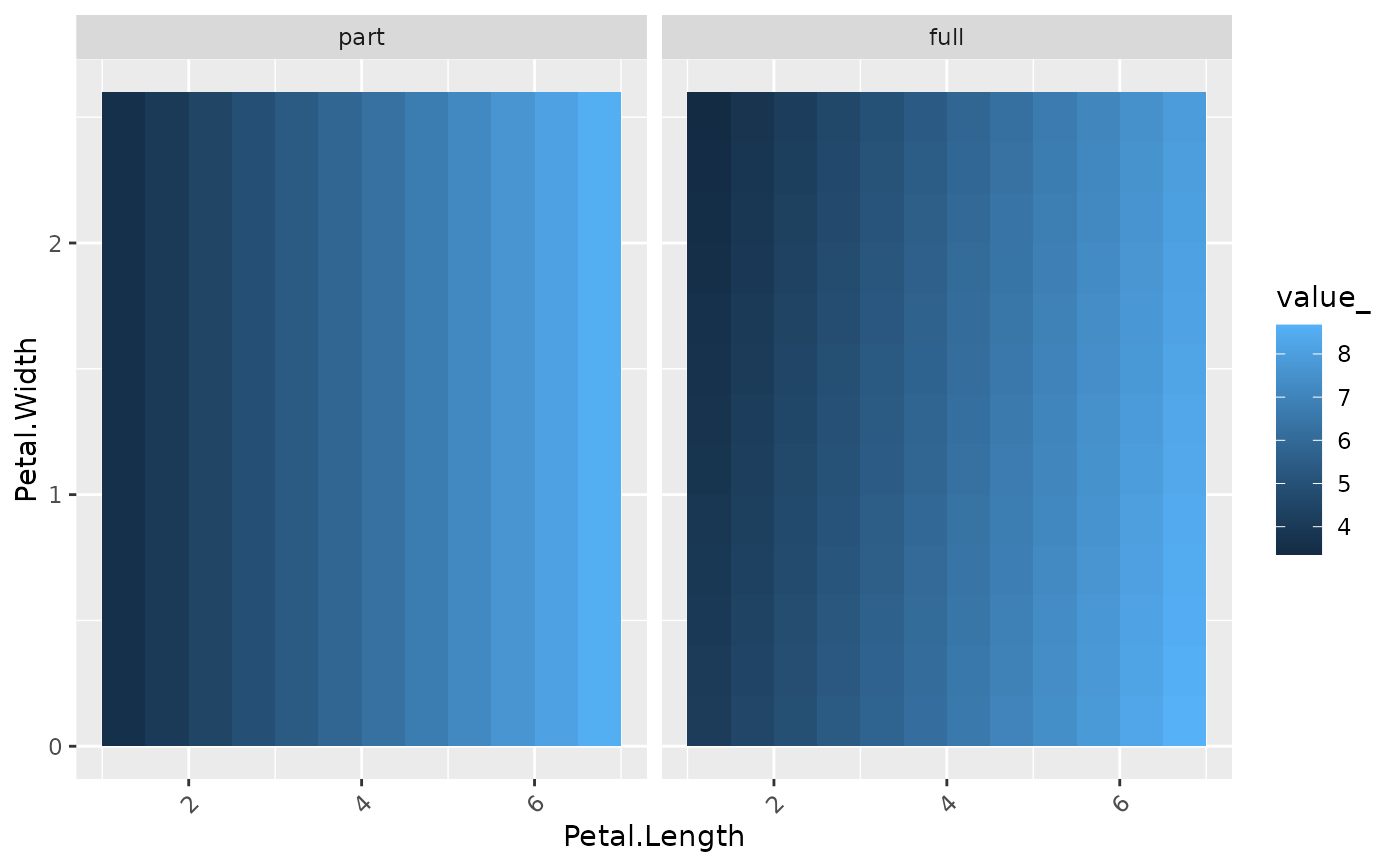
plot(light_profile2d(fl_part, v = c("Petal.Length", "Petal.Width")))
 # Second model includes Petal.Width
fit_full <- lm(Sepal.Length ~ ., data = iris)
fl_full <- flashlight(
model = fit_full, label = "full", data = iris, y = "Sepal.Length"
)
fls <- multiflashlight(list(fl_part, fl_full))
plot(light_profile2d(fls, v = c("Petal.Length", "Petal.Width")))
# Second model includes Petal.Width
fit_full <- lm(Sepal.Length ~ ., data = iris)
fl_full <- flashlight(
model = fit_full, label = "full", data = iris, y = "Sepal.Length"
)
fls <- multiflashlight(list(fl_part, fl_full))
plot(light_profile2d(fls, v = c("Petal.Length", "Petal.Width")))