READ THIS FIRST
The project has been integrated into the {kernelshap} package on CRAN. The code will run 1:1 from that library.
This repository is likely to be removed soon.
Overview
This package crunches exact permutation SHAP values of any model with up to 16 features. Multi-output regressions and models with case-weights are handled at no additional cost.
Later, we will extend the project to more features and approximate algorithms. The project emerged from a fork of our related project kernelshap.
The typical workflow to explain any model object:
-
Sample rows to explain: Sample 500 to 2000 rows
Xto be explained. If the training dataset is small, simply use the full training data for this purpose.Xshould only contain feature columns. -
Select background data: Permutation SHAP requires a representative background dataset
bg_Xto calculate marginal means. For this purpose, set aside 50 to 500 rows from the training data. If the training data is small, use the full training data. In cases with a natural “off” value (like MNIST digits), this can also be a single row with all values set to the off value. -
Crunch: Use
permshap(object, X, bg_X, ...)to calculate SHAP values. Runtime is proportional tonrow(X), while memory consumption scales linearly innrow(bg_X). - Analyze: Use {shapviz} to visualize the result.
Usage
Let’s model diamonds prices!
Linear regression
library(ggplot2)
library(permshap)
library(shapviz)
diamonds <- transform(
diamonds,
log_price = log(price),
log_carat = log(carat)
)
fit_lm <- lm(log_price ~ log_carat + clarity + color + cut, data = diamonds)
# 1) Sample rows to be explained
set.seed(10)
xvars <- c("log_carat", "clarity", "color", "cut")
X <- diamonds[sample(nrow(diamonds), 1000), xvars]
# 2) Select background data
bg_X <- diamonds[sample(nrow(diamonds), 200), ]
# 3) Crunch SHAP values for all 1000 rows of X (~10 seconds)
system.time(
shap_lm <- permshap(fit_lm, X, bg_X = bg_X)
)
shap_lm
# SHAP values of first 2 observations:
# log_carat clarity color cut
# [1,] 1.2692479 0.1081900 -0.07847065 0.004630899
# [2,] -0.4499226 -0.1111329 0.11832292 0.026503850
# 4) Analyze with shapviz >= 0.9.2 (Github install)
sv_lm <- shapviz(shap_lm)
sv_importance(sv_lm)
sv_dependence(sv_lm, "log_carat", color_var = NULL)We can also explain a specific prediction instead of the full model:
single_row <- diamonds[5000, xvars]
fit_lm |>
permshap(single_row, bg_X = bg_X)Random forest
We can use the same X and bg_X to inspect other models:
library(ranger)
fit_rf <- ranger(
log_price ~ log_carat + clarity + color + cut,
data = diamonds,
num.trees = 20,
seed = 20
)
shap_rf <- permshap(fit_rf, X, bg_X = bg_X)
shap_rf
# SHAP values of first 2 observations:
# log_carat clarity color cut
# [1,] 1.1986635 0.09557752 -0.1385312 0.001842753
# [2,] -0.4970758 -0.12034448 0.1051721 0.030014490
sv_rf <- shapviz(shap_rf)
sv_importance(sv_rf, kind = "bee", show_numbers = TRUE)
sv_dependence(sv_rf, "log_carat")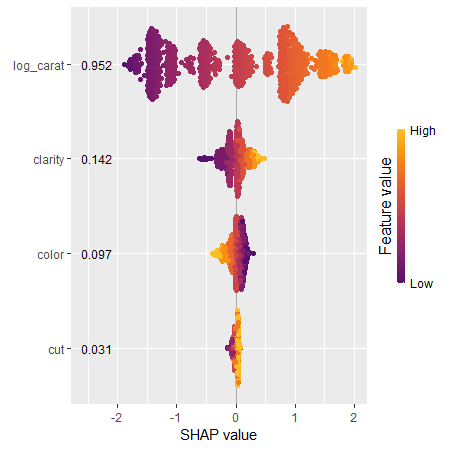
Deep neural net
Or a deep neural net (results not fully reproducible):
library(keras)
nn <- keras_model_sequential()
nn |>
layer_dense(units = 30, activation = "relu", input_shape = 4) |>
layer_dense(units = 15, activation = "relu") |>
layer_dense(units = 1)
nn |>
compile(optimizer = optimizer_adam(0.1), loss = "mse")
cb <- list(
callback_early_stopping(patience = 20),
callback_reduce_lr_on_plateau(patience = 5)
)
nn |>
fit(
x = data.matrix(diamonds[xvars]),
y = diamonds$log_price,
epochs = 100,
batch_size = 400,
validation_split = 0.2,
callbacks = cb
)
pred_fun <- function(mod, X)
predict(mod, data.matrix(X), batch_size = 10000, verbose = FALSE)
shap_nn <- permshap(nn, X, bg_X = bg_X, pred_fun = pred_fun)
sv_nn <- shapviz(shap_nn)
sv_importance(sv_nn, show_numbers = TRUE)
sv_dependence(sv_nn, "clarity")Parallel computing
Parallel computing is supported via foreach, at the price of losing the progress bar. Note that this does not work with Keras models (and some others).
Example: Linear regression continued
library(doFuture)
# Set up parallel backend
registerDoFuture()
plan(multisession, workers = 4) # Windows
# plan(multicore, workers = 4) # Linux, macOS, Solaris
# ~4 seconds
system.time(
s <- permshap(fit_lm, X, bg_X = bg_X, parallel = TRUE)
)Example: Parallel GAM
On Windows, sometimes not all packages or global objects are passed to the parallel sessions. In this case, the necessary instructions to foreach can be specified through a named list via parallel_args, see the following example:
library(mgcv)
fit_gam <- gam(log_price ~ s(log_carat) + clarity + color + cut, data = diamonds)
system.time(
shap_gam <- permshap(
fit_gam,
X,
bg_X = bg_X,
parallel = TRUE,
parallel_args = list(.packages = "mgcv")
)
)
shap_gam
# SHAP values of first 2 observations:
# log_carat clarity color cut
# [1,] 1.2714988 0.1115546 -0.08454955 0.003220451
# [2,] -0.5153642 -0.1080045 0.11967804 0.031341595Meta-learning packages
Here, we provide some working examples for “tidymodels”, “caret”, and “mlr3”.
tidymodels
library(tidymodels)
library(permshap)
iris_recipe <- iris %>%
recipe(Sepal.Length ~ .)
reg <- linear_reg() %>%
set_engine("lm")
iris_wf <- workflow() %>%
add_recipe(iris_recipe) %>%
add_model(reg)
fit <- iris_wf %>%
fit(iris)
ps <- permshap(fit, iris[, -1], bg_X = iris)
psmlr3
library(permshap)
library(mlr3)
library(mlr3learners)
# Regression
mlr_tasks$get("iris")
task_iris <- TaskRegr$new(id = "reg", backend = iris, target = "Sepal.Length")
fit_lm <- lrn("regr.lm")
fit_lm$train(task_iris)
s <- permshap(fit_lm, iris[-1], bg_X = iris)
s
# Probabilistic classification
task_iris <- TaskClassif$new(id = "class", backend = iris, target = "Species")
fit_rf <- lrn("classif.ranger", predict_type = "prob", num.trees = 50)
fit_rf$train(task_iris)
s <- permshap(fit_rf, X = iris[-5], bg_X = iris)
s
